- Feb. 11, 2019
- BugSpeaks
- On Microbiome
The microbiome role in human health and diseases
In recent years, studies on the native microflora (microbiota) of the host environment has profoundly renewed our knowledge of human biology, health and diseases. The term, microbiome is generally referred to the pool of genes present within a community of microbes including bacteria, fungi, archaea, protozoa, and viruses found over and inside the body. While, studies aiming to describe the occurrence and function of uncultured microbiota are frequently stated collectively as metagenomics. The life style of the host including diet habits, medical interventions, environment, and disease conditions can lead to variations in the microbiome. The structural and functional diversity of microbes allow them to survive symbiotically for a long-term in hosts. The microbiome contributes to the vital functions of the body such as metabolism, immunologic activity, energy homeostasis, gut epithelial health and neurodevelopment of the gut brain axis. They influence or mediate the process of digestion in gut, regulate immune system, protect against other pathogenic bacteria and secrete vitamins including Thiamine, Vitamins B12, Vitamin K, etc.. Overall, microbiota improves human health and helps to combat diseases. For instance, gut microbes make drugs to work better. On the other hand, they play a significant role in causing several disease including inflammatory bowel disease, autism, Parkinson's disease, type 2 diabetes, cancer, and depression. In addition, they can affect therapeutic approaches by reducing drug’s effectiveness.
The advent of next generation sequencing (NGS) technologies, metagenomics, transcriptomics, metabolomics, and analytical methods combined with the latest analytical software and the unified databases on experimental results have revolutionized the emergence of a new field, the microbiomics. It aims to identify the microbiota, genome DNA analysis, elucidate the microbiome and host interactions, and determine its role in disease pathogenicity. Basically, these approaches are used to amplify and sequence the specific microbial DNA segments followed by integrating computational analysis to identify and diversify microbes on the basis of sequence similarity and comparing with reference genomic databases of microbes. In specific, 16S rDNA (ribosome DNA) is used as the target for sequencing bacterial genome as this region is highly conserved with hypervariability and vary amongst species. In recent times, whole-genome shotgun sequencing (WGS) is the choice of analysis. This is because, WGS permits taxonomic scrutiny, investigations on the microbial gene inventories, and documents non-bacterial microorganisms that are omitted from 16S rDNA sequencing (Figure 1). Initiatives of various projects such as the Metagenomics of the Human Intestinal Tract (MetaHIT), Human Microbiome Project (HMP) have quickly enriched the microbial genomic data. This mammoth and wealthy data rapidly led to overcome computational bottlenecks and driven to develop tools such as Quantitative Insights into Microbial Ecology (QIIME) and Visualization and Analysis of Microbial Population Structures (VAMPS) which can handle big volumes of genomic data and can function in different settings ranging from supercomputers to laptops. Estimates suggest that the human body is associated with approximately 2-20 million number of microbial genes. The studies have reported the ratio of microbial cells to human cells to be 10:1. This suggest the profound impact of microbes and their genomic functions on human biology. Thus, exploration of their genomic DNA can offer exciting prospects for developing unique therapies against many chronic diseases.
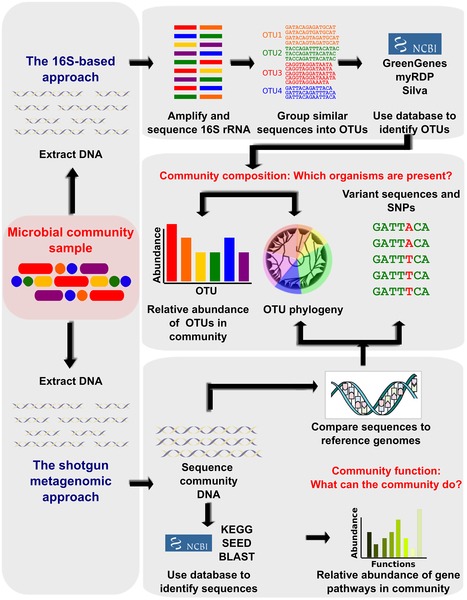
Figure 1. Bioinformatic methods for functional metagenomics.
(Source: Adopted from Morgan and Huttenhower (2012); https://doi.org/10.1371/journal.pcbi.1002808)
The microbes living within us are not invaders, but helpful colonizers and play an essential role in human development, nutrition and immunity. Several autoimmune diseases including diabetes, muscular dystrophy, rheumatoid arthritis, and multiple sclerosis, and fibromyalgia are linked with dysfunctioning of the microbiome. The higher number of Lactobacillus counts in pregnancy is linked to protect the developing infant from allergic reactions. Also, studies have suggested that Lactobacillus-dominated populations are protected from bacterial vaginosis. A failure in the vaginal microbiome may increase the risk of pre-term birth. Thus, the mother’s microbiome may influence her children’s health. Likewise, disease-causing microorganisms add over time, and change the gene action and metabolic functions. This may lead to irregular immune responses against body’s own constituents and tissues and results in autoimmune diseases. Overall, varied microbiome of an individual can make them susceptible to infectious diseases and lead to Crohn’s disease, irritable bowel syndrome and other chronic disorders of the intestine. Some microbial communities regulate the response of a person treated with a drug. When, a broad-range antimicrobials are taken both good and bad bacteria are killed. This might lead to the antibiotic resistance crisis. Therefore, there is a need to accurately identify pathogenic microbes so as to know their role in contributing diseases and strategize treatments which can selectively kill only pathogenic bacteria. Therefore, mapping the human microbiome is necessary to discover more microbial species and their genes that are associated with definite human health conditions. The microbiome varies depending on the population, race, geography, and diet habits. The microbiomics can help clinicians to prescribe the right drugs and diet to manage diseases effectively. The addition of probiotics is one of the nutritional supplementation to improve the microbiota of the gut. However, the application of microbiome in therapies is challenging because microbiomes are distinctive to every single person and vary over time. Thus, it complicates the use of the microbiome for both diagnosis and to design a therapy. At present, research studies are aimed to investigate on the following aspects:
- Nutrition in diet affecting the microbiome.
- The microbiome affecting immune response and contributing to diseases.
- Antibiotics and the microbiome interactions to overcome drug-resistance.
- The microbial response to various drugs.
- Altering the microbiome to improve health conditions.
The microbiome is very diverse, and complex and many fundamental questions remain unanswered. Improved computational approaches for the metagenome assembly/analysis are the urgent need in this regard. Moreover, their relations to metabolism, epigenetics, host genetics, and immunology can facilitate to unravel their complex relationships with human health and diseases. This can be better exploited for curing several chronic diseases. Some of the ethical concerns of the microbiomic research include; data being obtained from a typical sample of the population, informing about the consent to samples collection, data sharing, protecting privacy, invasiveness of taking the samples, etc. In future, investigations should aim to explain the mechanisms of interactions between the microbiome and host, decipher the microbiome growth during host growth and development, its impact on health conditions, elucidate the role of its pathogenicity, and measure the feasibility of diagnostic examinations, and target the therapies against human diseases.