- Feb. 11, 2019
- BugSpeaks
- Others
Antibiotics influence on the gut microbiome functions
The current research findings have witnessed a revolution in the biological sciences and, unambiguously, in human health care. In this regard, ‘omics’ approaches, such as genomics, proteomics and metabolomics have helped us to unlock the paradigm of microbiome role in improving human health. A balanced maintenance of a good microbiota in our body significantly promotes our health conditions and keep away many metabolic diseases.
The microbiome usually exists in our body through an intimate symbiotic relationship, and the status of microbial flora can be an indicator of a good health. However, the microbiota is constantly being exposed to several factors that might impact on it vigorously. The microbial composition and its alterations vary from one individual to another, and this mainly depends on the microbiota stability of an individual and the nature of a disturbing factor. Under some circumstances, when the environmental or other disturbing factors are very strong, they can induce stress in our body sites, especially the gut location leading to imbalance in the taxonomic configuration of the microbiota, and their functions (a dysbiotic state). For instance, the composition of microbiome is rapidly altered when exposed to antibiotics, and may cause many possible direct or indirect health effects (Figure 1).
Antibiotics are known to kill pathogens. They are classified into natural, semisynthetic and synthetic based on their production mode and origin. For example, the fungus, Penicillium notatum producing penicillins is the base for most of the beta-lactam antibiotics. The treatment of diseases using antibiotics must be considered with a proper dosage. However, when antibiotics are consumed wrongly (not specific to the pathogen) or at high concentrations, i.e., they produce co-lateral effects in our microbiota. It has been reported that broad-spectrum antibiotics affect the richness of bacteria (up to 30%) in the gut microbial community, and cause a significant dysbiotic state. Indeed, the microbiota alterations due to antibiotics may remain for long periods, even up to a few years. Likewise, in infants, it has been evidenced that early exposure to antibiotics significantly reduces the microbial diversity of the infants’ microbiota, with weakening of Bifidobacterium and marked increases of Proteobacteria. Such effects of antibiotics on the gut microbiome have been investigated thoroughly in recent times with the availability of a variety of ‘omic’ approaches. The changes are mainly identified by 16S ribosomal RNA gene sequences analysis and shotgun sequence datasets evaluation. These researches have revealed that, in addition of altering the composition of microbiota, antibiotics upset the gene expression, biomolecules function and inclusive metabolic activities of the gut microbiota. Antibiotics swiftly modify the physiological state and functions of the gut microbiota.
The antibiotic-induced gut microbiome alter the immune and metabolic health of an individual. An immediate threat of an altered gut microbiome is the augmented vulnerability to intestinal infections, which could be because of the newly assimilated pathogens or sudden over-growth and pathogenicity by the already existing opportunistic microbes in the microbial flora. For example, antibiotic-associated diarrhea caused by the nosocomial pathogens, such as Staphylococcus aureus, Klebsiella pneumoniae, and Clostridium difficile is a common incidence. Studies have provided enough evidences to suggest that a chronic infection with C. difficile emerges with a loss of intestinal microbial diversity due to antibiotics. In a similar way, bloodstream infections in immunocompromised patients are increasing due to antibiotics treatments. Researchers have shown that bloodstream infections treated with vancomycin (an antibiotic) set a stage for the intestinal outgrowth of vancomycin-resistant Enterococcus. Studies have revealed that bacteria belonging to a limited set of phyla and genera are strongly affected by antibiotics. Fluoroquinolones, being the most widely prescribed antibiotics are reported to influence our microbiota to a greater extent with the modifications in about 32 major microbial groups. Hence, it has led to the increased resistance worldwide. The major phyla effected include Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria. In the major genera, Bifidobacterium, Escherichia, Bacteroides, Ruminococcus, Enterobacter, Faecalibacterium, Bacteroides, Lactobacillus, Eubacterium and Faecalibacterium are highly influenced by antibiotics. The effects of usages of a single antibiotic or a cocktail of antibiotics significantly disturb the microbial community. For example, the treatment with ampicillin alter only the bacteria belonging to the genera Klebsiella, Streptococcus, Bacillus, and Salmonella. Some antibiotics reduce the richness of bacteria community, which is beneficial for human health. For instance, bacteria belonging to the genus, Faecalibacterium, Bifidobacterium, and Blautia to name a few.
Few studies have investigated the influence of antibiotics on our microbes, especially gut microbiome. They suggested the role of microbial diversity and activity, expression of genes, levels of proteins or metabolites produced by microbes in their community. Antibiotics damage and/or destroy bacterial cells and therefore decreases enzymatic activities. However, in the process of antibiotic medications, antibiotic-vulnerable bacteria might be changed into resistant bacteria.
During the treatment course of antibiotics, alterations in specific enzymatic activities have been witnessed, for example the hydrolysis of dietary polysaccharides. Largely, treatments with ampicillin, cefazolin, and sulbactam are reported to alter the activity level of glycoside hydrolases and favor the rapid or disturbed absorption of carbohydrates leading to type-2 diabetes and obesity. Likewise, antibiotics treatment can cause alterations in the microbial metabolite production. In addition, antibiotics bring changes in the fluxes of genes and metabolites and the activities they mediate. A study has detected over 87% of all fecal metabolites in abundance due to streptomycin treatment. This suggests that antibiotics impacts the metabolite fluxes to a different extent. As stated by Ferrer et al. (2017), a comparative ‘omic’ investigation of microbial communities in fecal samples taken at multiple time points from an individual (with a bacterial infection) subjected to β-lactam therapy of ampicillin, sulbactam and cefazolin has revealed antibiotic-associated imbalances in long linear and branched, saturated and unsaturated fatty acids, branched chain amino acids, cholesterol derivatives, vitamins, polyols, sugars, short peptides and polyamines. Likewise, antibiotic intervention during pregnancy causes variations or copiousness of short-chain fatty acids, mostly, propionate, acetate, and butyrate, which are linked to the delivery of premature babies.
Recent years have been noticed with a rapid rise in the cases of multidrug-resistant microorganisms worldwide. This has become a great threat to the use of antibiotics for treating several microbial diseases. The resistance catastrophe is mainly attributed to the misuse and overuse of antibiotics/drugs. The effects of over-use of antibiotics are not only observed in pathogenic gut bacteria, but also in symbiotic healthy microbiota. In this regard, a preventive measures are very necessary to antibiotic-based treatment tactics that do not disturb or unbalance our inhabitant microbial flora, especially good gut bacteria having a dynamic role in health promotion.
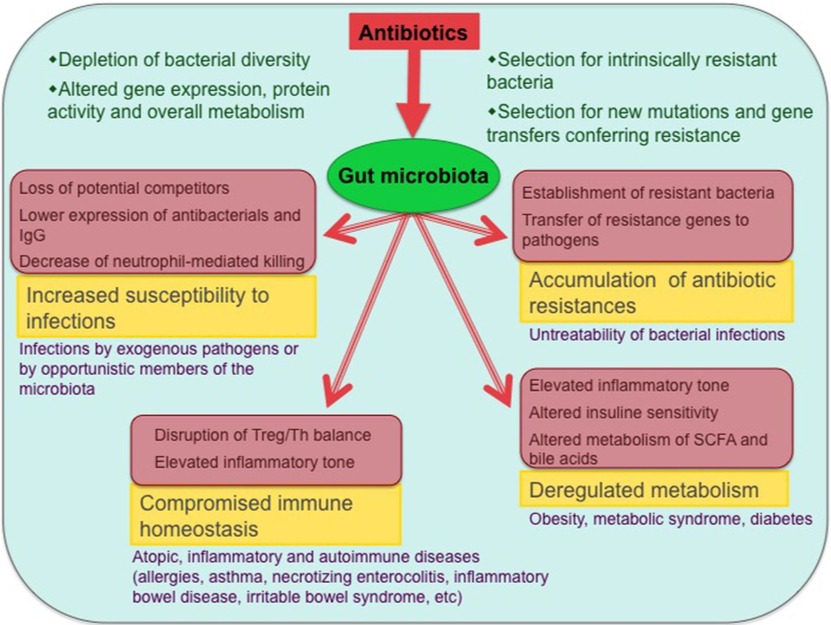
Figure-1: Antibiotic effects on the gut microbiota and associated health problems. The main biological consequences of antibiotic-induced dysbioses and the potential diseases that can ensue from them are shown (only diseases with published evidence of association with antibiotic exposure are included). Involved mechanisms are shown in pink-shaded boxes.
(Source: Francino, 2016; https://doi.org/10.3389/fmicb.2015.01543).
References:
- Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Frontiers in microbiology. 2016 Jan 12;6:1543.
- Ferrer M, Méndez-García C, Rojo D, Barbas C, Moya A. Antibiotic use and microbiome function. Biochemical pharmacology. 2017 Jun 15;134:114-26.