- Feb. 11, 2019
- BugSpeaks
- Others
The human gut microbiome and microbiomics: progress towards personalized healthcare
The human body comprises 10-100 trillions of microbial cells, including archaea, bacteria, viruses, eukaryotes. The microbiome includes the combined genetic material of all microbes in a specific environment i.e., microbiota. It plays a significant role in improving human health and preventing diseases in many ways, though their mechanisms are yet to be probed in detail. The occurrence and function of microbiota differ depending on several factors, such as ages, sexes, races, diets, and change in the locations of the host. The microbiome profoundly restores normal phenotypes in the host. The past few years have investigated the microbiome in detail, and the results have tremendously reshaped our knowledge of human biology. Some of the innovative insights range from the understanding of how microbial cells intervene digestion activities, disease initiation, and progressions (e.g., inflammatory bowel disease) to unforeseen links with autism, depression, anxiety/mood disorders, Alzheimer's, Parkinson's disease, etc.
So far, studies have discovered around 10,000 microbial species (mainly bacteria) in the human body and mostly, their occurrence is predominant in the gut. Interestingly, the gut microbiota contains about 150 times more number of genes than that are observed in the whole human genome. Thus, the gut microbiome is sometimes recognized as an ‘essential organ’ because it produces products, it responds to the environment, and interacts with other systems. It is believed that the impact of these bacterial genomic DNA on our health might be more than our own genomic DNA. Likewise, it has been realized that the microbiota of gut and brain are intricately associated. There is an exchange of messengers/signals between the brain and gut microbiota. The entire body cells are influenced by the microbiome in one or the other way. In specific, it affects our mood, metabolism, libido, immune system, sensitivity, and even the clarity of your thoughts. Scientific evidences have shown that human health and diseases are dictated by what goes on in the microbiome of the gut. Diseases such as inflammatory bowel disease (IBD), obesity, diabetes, cardiovascular diseases, atherosclerosis, nonalcoholic fatty liver disease (NAFLD), alcoholic liver disease (ALD), cirrhosis, and colorectal and liver cancer have been connected to the activities of the human microbiota (Figure 1).
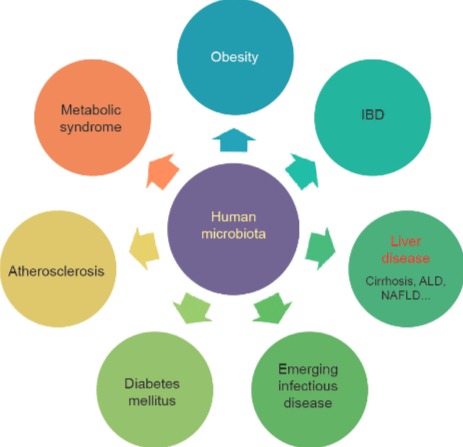
Figure 1. Human microbial symbiosis has a close relationship with diseases of different systems (Adopted from Wang et al. 2017; https://doi.org/10.1016/J.ENG.2017.01.008).
Overall, the diversity in microbiome can lead to both health and illness. Commensal microbes colonize the host soon after their birth. This microbiota community gradually develops with the host growth depending on varying ecosystems. Over time, these host-microbe interactions lead to a beneficial association, and symbiotically offer many advantages. For example, improvements in digestion, supply of essential nutrients, metabolizing indigestible compounds, and to shield against the invading pathogens. Knowing about the microbiota can help in preventing possible diseases in humans. However, their complex nature makes it difficult to study them. The standard microbial culture methods may not be sufficient enough to isolate and explore all microbes present, because some may not grow or require special growth conditions that are yet to be standardized. Later, culture independent methods for evaluating microbes was developed based on the targeted sequencing of 16S and 5S ribosomal RNA (rRNA) genes which showed differences for each species and effective in identifying organisms. Over the last decade, human microbiome researches have been transformed by using next-generation sequencing (NGS) technologies. These high-throughput sequencing allow us to study complex bacterial systems without the requirement of cloning individual genes. The targeted sequencing of 16S rRNA, 18S rRNA, 28S rRNA genes, and internal transcribed spacer (ITS) regions, and whole microbial genome of microbial communities all at once (metagenomics) is very useful in the identification of microbes and to track the functional activity of the entire community. The increasing number of such metagenomic studies of microbiomes in humans is a new hope of its potential benefits in the clinical practice.
Recent studies have witnessed the occurrence of human genetic variations as influenced by interpersonal dissimilarities in microbiomes. Thus, human genes might influence health by promoting beneficial microbiomes. The heritability study of the gut microbiota has revealed that subsets of microorganism’s abundance are genetically determined by the host. For example, a genetic change enables lactase persistence in adulthood. Interestingly, the genome-wide association studies (GWASs) of the microbiome has revealed that there is a relationship between the host genotype, consumption of milk, and gut beneficial bacteria (Bifidobacteria). It has been noticed that a single nucleotide polymorphism (SNPs) near the LCT gene on chromosome 2 and Bifidobacterium in the fecal microbiota has a close association. Bifidobacteria utilizes the lactose of milk as an energy source. This depends on the genotype of the host and diet interaction (Figure 2).
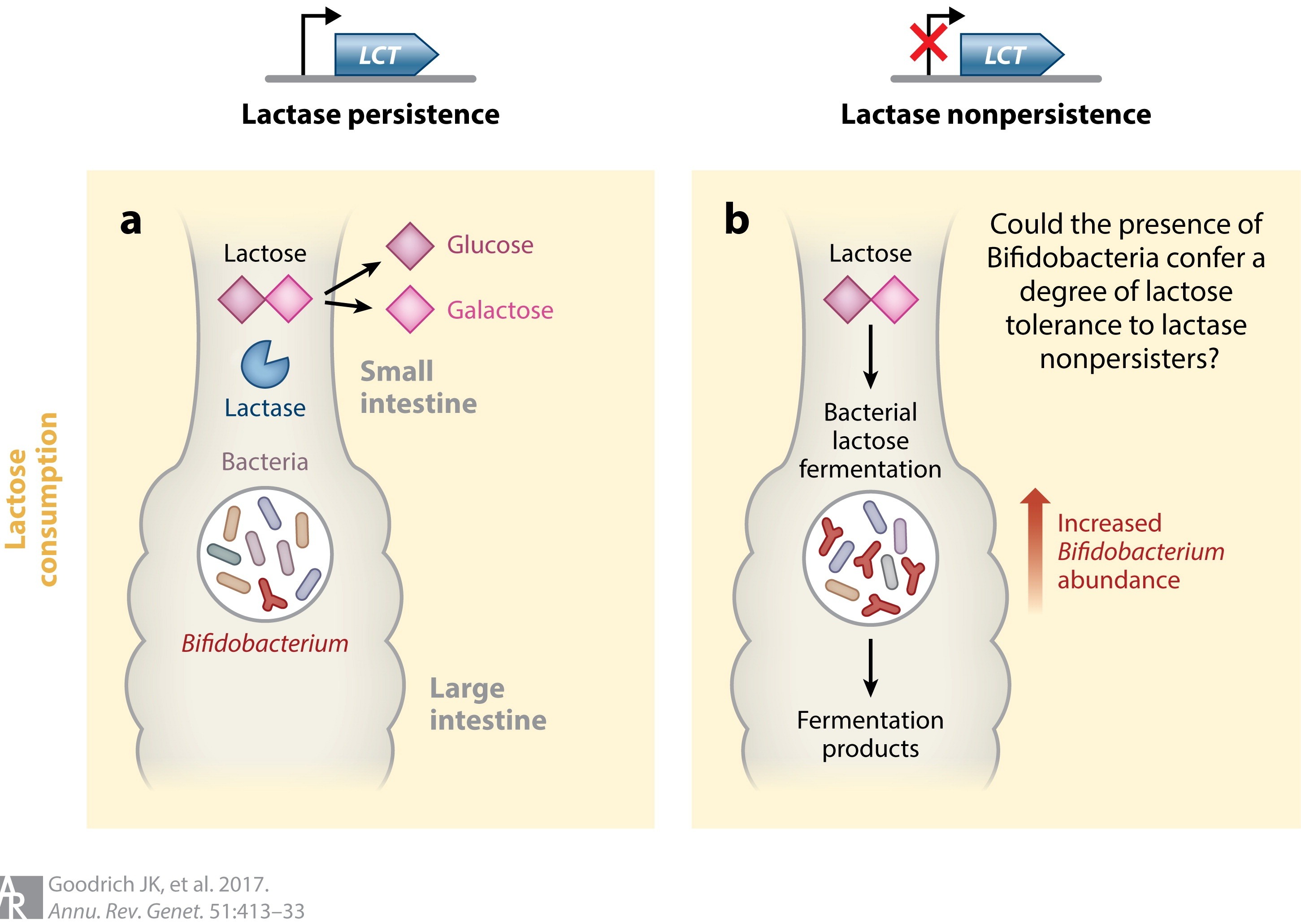
Figure 2: Interaction between the human lactase nonpersister genotype, Bifidobacterium abundance, and lactose in the host’s diet. Genetic variants near the LCT gene are associated with lactase persistence and in several microbiome GWASs were recently associated with Bifidobacterium relative abundance in fecal samples. The association may function according to the following scenario: (a) If an individual is a lactase persister and consumes lactose, the lactose is typically broken down into glucose and galactose in the small intestine by host lactase. (b) However, if the individual is a lactase nonpersister and consumes lactose, it travels to the large intestine, where it is fermented by lactose-utilizing bacteria, which includes Bifidobacterium. If Bifidobacteria are present, then the presence of lactose promotes their abundance. In individuals who do not consume lactose, Bifidobacterium abundance remains unaffected by their lactase persistence status (not shown). (Adopted from Goodrich et al. 2016; https://doi.org/10.1016/j.cell.2014.06.037).
Likewise, GWASs has revealed that immunity related genes are influenced by the gut microbiome. Evidences suggest that genetic variations might act on digestive tract tissues to disturb the composition of the gut microbiome. Further, genetic variants in the genes, PLD1 and LINGO2 are associated with obesity. Based on the analysis of 16S transcripts, the phylogeny of microbial community has been revealed. The most active microbiota included Prevotellaceae, Lachnospiraceae, Bacteroidaceae, Ruminococcaceae, and Rickenellaceae. RNAseq analysis of bacteriophage populations in the periodontal microbiota showed the abundance of oral phages highly expressed in persons having good periodontal health. More recently, 16S rRNA gene-based approach has confirmed the predominant occurrence of Corynebacterium spp., Staphylococcus spp. and Propionibacterium spp. on healthy human skin.
Thus, knowledge on the microbiomics help to improve human health conditions. A defined diet can influence on the microbiome and restore health. Likewise, a proper use of chemicals/drugs in disease management is another factor to be considered. For example, both foods and drugs are used in treating type 2 diabetes. It is generally advised to avoid consuming simple carbohydrates to control blood sugar. Nevertheless, there exist inter-individual glycemic variations in response to diet, and some of which can be described on the basis of the individual’s microbiome. Similarly, the human microbiome plays a significant role in genetic diversity, modify disease conditions, impart immunity, provide nutrients, influences assimilation, and modulates drug interactions. Thus, intake of probiotics or beneficial bacteria (Lactobacillus, Bifidobacterium, Faecalibacterium prausnitzii, Akkermansia muciniphila, Pediococcus pentosaceus, etc.) may be useful in preventing or treating certain diseases, though most of them cannot be cultured presently. As a diagnostic tool, microbiomics is employed for detecting and measuring the levels of biomarkers i.e., cholesterol, cortisol, glucose, and creatine kinease occurring in blood which are known to indicate about diseases or infections. These novel diagnostic methods will allow clinicians to prescribe right drugs or diet to restore patient’s health.
Future microbiome investigations should involve many more new techniques to predict the functions of microbiotas, their crucial role in human development, and the mechanisms of various diseases, such as metabolic diseases, liver diseases, cancers, psychiatric diseases, etc. In this regard, many new-startups and biopharmaceutical companies, including Luecine Rich Bio Pvt. Ltd are exploring the field of microbiomics to develop novel therapeutics, diagnostic platforms and to provide clinical genomic services. A detailed data on this metagenomics might be very useful in the microbiome-based diagnosis of diseases and for the development of novel treatment strategies in the forthcoming personalized medicine era.